My research area is computational biology. I develop and use computational and statistical methods to interpret complex biological datasets. In view of the recent progress in next-generation sequencing (NGS) technology, which is driving rapid accumulation of biological data, robust analysis and interpretation of this data is of crucial importance. My research aims to address this ‘big data’ challenge. Central to my research are the questions as to:
1. How despite having same genetic code a huge diversity of cell arises?
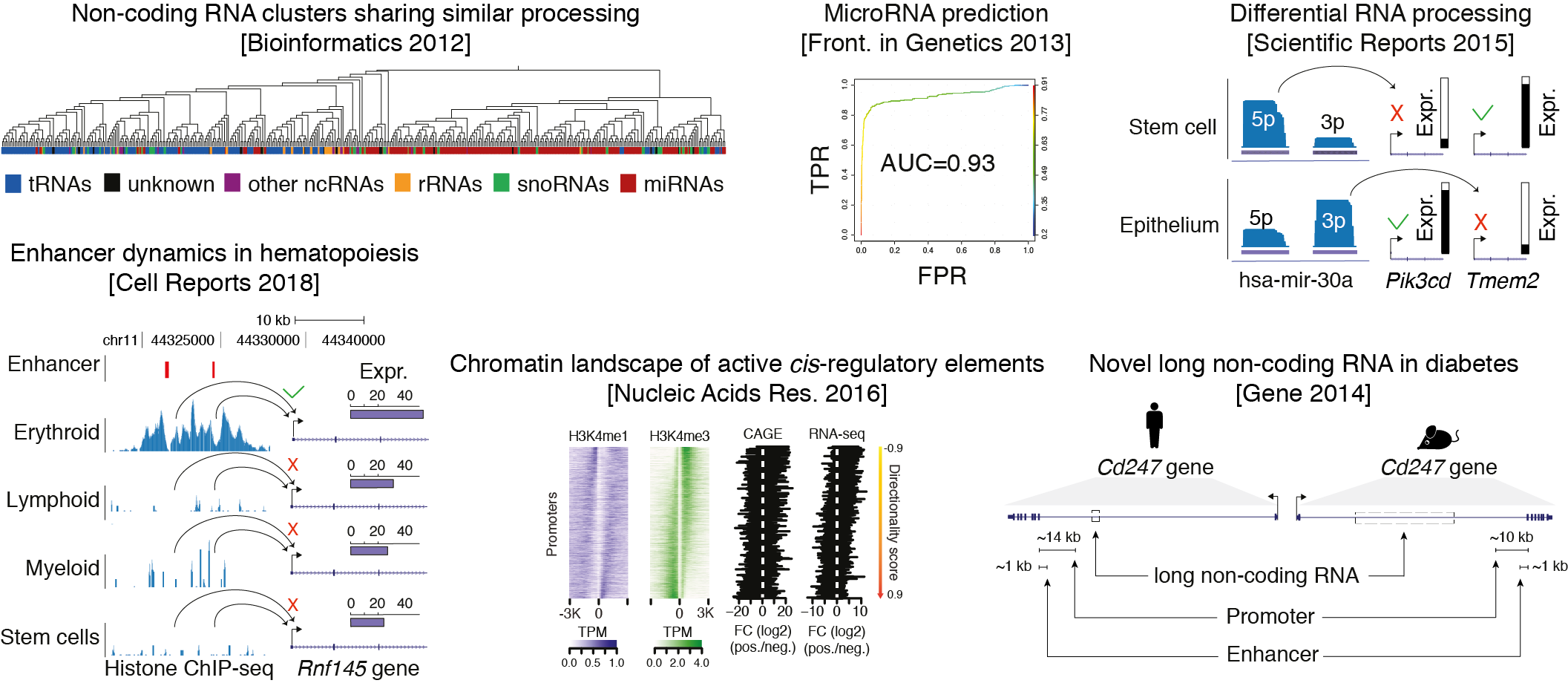
2. How do the non-coding RNA (RNA-seq), transcription factor and chromatin (ChIP-seq) landscape define the identity of a cell?
3. How do alterations in this landscape drive cellular differentiation, and if aberrant to the formation of cancer cells?
These are fundamental questions, an exploration of which can lead to better prognosis of disease and personalized medicine. However, they also raise several challenges due to non-coherent nature of the data and the inherently interdisciplinary nature of the problems.
My goal is to address these questions by proposing new computational methods that use standardized NGS data formats and robust statistics to capture patterns unique for a particular cell state or disease condition, and by maintaining collaborations with biologists for experimental follow-up and validation.
My research statement is available here.
My teaching statement is available here.